Nucleophilicity Model
- Prediction methods: Extra Trees and rSPOC (reactivity SPOC) descriptors with MAE=1.00 and r2=0.96 (90:10 train test split).
- Dataset: The nucleophilicity dataset contains 1115 experimental N parameters. The experimental data comes from the Mayr’s Database of Reactivity Parameters, for more details please refer to Mayr's Database.
Electrophilicity Model
- Prediction methods: Extra Trees and SPOC descriptors with MAE=1.49 and r2=0.92 (90:10 train test split).
- Dataset: The electrophilicity dataset contains 285 experimental E parameters. The experimental data comes from the Mayr's Database of Reactivity Parameters, for more details please refer to Mayr's Database.
Notes
- The dataset was refined during model construction by double-checking the outliers. For the N-dataset, the nucleophilicities of transition-metal complex, solvent mixtures, or those of multi-site reactant, P-nucleophiles and H-P nucleophiles were removed due to their scarcity. For E-dataset, we mainly focused on carbon-based electrophiles and those hetero-atom-centered or organometallic electrophiles were excluded.
- For SPOC descroptors, please refer to our previous work about pKa prediction and SPOC.
- Abbreviations of solvents: DCM - dichloromethane, MeCN - Acetonitrile, DMSO - Dimethyl sulfoxide.
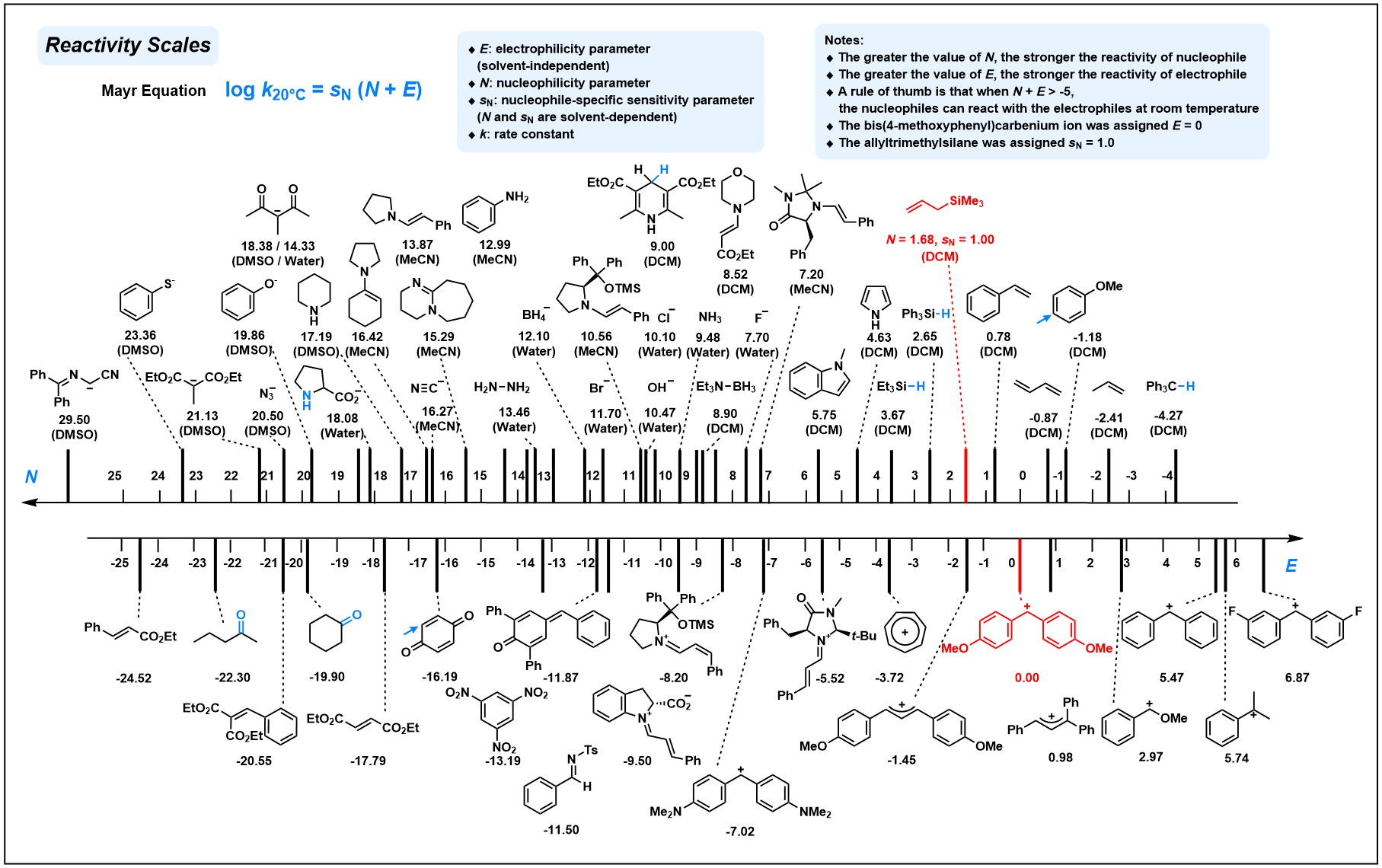
Extracted from Mayr's Reactivity Scales Poster. For complete reactivity scales, please refer to: www.cup.lmu.de/oc/mayr/MayrPoster.html